11. AIH
Christian P. Strassburg
(This chapter corresponds to the tenth edition from 2020.)
Autoimmune hepatitis (AIH)
Autoimmune hepatitis (AIH) is a chronic inflammatory disease, in which a loss of tolerance against hepatic tissue is presumed. Autoimmune hepatitis (AIH) was first described as a form of chronic hepatitis in young women showing jaundice, elevated gamma globulins and amenorrhoea, which eventually led to liver cirrhosis (Waldenström 1950). A beneficial effect of steroids was described in the reported patient cohort and thus the groundwork was laid for the first chronic liver disease found to be curable by drug therapy. AIH was later recognised in combination with other extrahepatic autoimmune syndromes, and the presence of antinuclear antibodies (ANA) led to the term lupoid hepatitis (Mackay 1956). Systematic evaluations of the cellular and molecular immunopathology, of the clinical symptoms and of laboratory features has subsequently led to the establishment of autoimmune hepatitis as a clinical entity on its own, which is serologically heterogeneous, treated by an immunosuppressive therapeutic strategy (Strassburg 2000). An established (Alvarez 1999a) and recently simplified (Hennes 2008b) revised scoring system allows for a reproducible and standardised approach to diagnosing AIH in a scientific context but has limitations in everyday diagnostic applications. The use and interpretation of seroimmunological and molecular biological tests permits a precise discrimination of autoimmune hepatitis from other etiologies of chronic hepatitis, in particular from chronic viral infection as the most common cause of chronic hepatitis worldwide (Strassburg 2002). Today, AIH is a treatable chronic liver disease in the majority of cases. Much of the same initial treatment strategies of immunosuppression still represent the standard of care. The largest challenge regarding treatment is the timely establishment of the correct diagnosis.
Definition and diagnosis of autoimmune hepatitis
In 1992, an international panel met in Brighton, UK, to establish diagnostic criteria for AIH because it was recognised that several features including histological changes and clinical presentation are also prevalent in other chronic liver disorders (Johnson 1993). In this and in a revised report the group noted that there is no single test for the diagnosis of AIH. In contrast, a set of diagnostic criteria was suggested in the form of a scoring system designed to classify patients as having probable or definite AIH (Table 1). According to this approach the diagnosis relies on a combination of indicative features of AIH and the exclusion of other causes of chronic liver diseases. AIH predominantly affects women of any age, and is characterised by a marked elevation of serum globulins, in particular gamma globulins, and circulating autoantibodies. It should be noted that AIH regularly affects individuals older than 40 but should be considered in all age groups (Strassburg 2006). The clinical appearance ranges from an absence of symptoms to a severe or fulminant presentation (Stravitz 2011) and responds to immunosuppressive treatment in most cases. An association with extrahepatic autoimmune diseases such as rheumatoid arthritis, autoimmune thyroiditis, ulcerative colitis and diabetes mellitus and a family history of autoimmune or allergic disorders has been reported (Strassburg 1995).
Autoantibodies are one of the distinguishing features of AIH. The discovery of autoantibodies directed against different cellular targets including endoplasmatic reticulum membrane proteins, nuclear antigens and cytosolic antigens has led to a suggested subclassification of AIH based upon the presence of three specific autoantibody profiles. According to this approach, AIH type 1 is characterised by the presence of antinuclear antibodies (ANA) and/or anti-smooth muscle antibodies (SMA) directed predominantly against smooth muscle actin. AIH type 2 is characterised by anti-liver/kidney microsomal autoantibodies (LKM-1) directed against cytochrome P450 CYP2D6 (Manns 1989, Manns 1991) (Figure 1) and with lower frequency against UDP-glucuronosyltransferases (UGT) (Strassburg 1996). AIH type 3 (Manns 1987, Stechemesser 1993) is characterised by autoantibodies against a soluble liver antigen (SLA/LP) identified as UGA suppressor serine tRNA-protein complex (Gelpi 1992, Wies 2000, Volkmann 2001, Volkmann 2010). However, this serological heterogeneity does not influence the decision of whom to treat or of what strategy to employ.
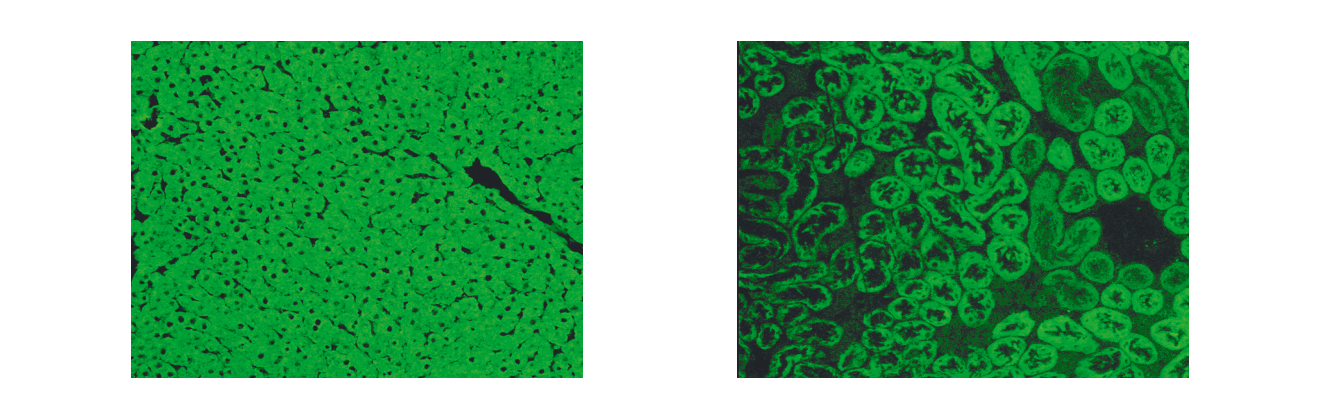 Figure 1. Indirect immunofluorescence showing LKM-1 autoantibodies on rat kidney and liver cryostat sections. Serum of a patient with autoimmune hepatitis type 2. A) Using rat hepatic cryostat sections a homogeneous cellular immunofluorescence staining is visualised excluding the hepatocellular nuclei (LKM-1). B) Typical indirect immunofluorescence pattern of LKM-1 autoantibodies detecting the proximal (cortical) renal tubules but excluding the distal tubules located in the renal medulla, which corresponds to the tissue expression pattern of the autoantigen CYP2D6
Figure 1. Indirect immunofluorescence showing LKM-1 autoantibodies on rat kidney and liver cryostat sections. Serum of a patient with autoimmune hepatitis type 2. A) Using rat hepatic cryostat sections a homogeneous cellular immunofluorescence staining is visualised excluding the hepatocellular nuclei (LKM-1). B) Typical indirect immunofluorescence pattern of LKM-1 autoantibodies detecting the proximal (cortical) renal tubules but excluding the distal tubules located in the renal medulla, which corresponds to the tissue expression pattern of the autoantigen CYP2D6
Although the histological appearance of AIH is characteristic, there is no specific histological feature that can be used to prove the diagnosis (Dienes 1989). Percutaneous liver biopsy is recommended initially for grading and staging (EASL 2015), as well as for therapeutic monitoring when this is considered necessary for therapeutic planning. Histological features usually include periportal hepatitis with lymphocytic infiltrates, plasma cells, and piecemeal necrosis. With advancing disease, bridging necrosis, panlobular and multilobular necrosis may occur and ultimately lead to cirrhosis. A lobular hepatitis can be present, but is only indicative of AIH in the absence of copper deposits or biliary inflammation. However, biliary involvement does not rule out AIH. The presence of granulomas and iron deposits argue against AIH.
Viral hepatitis should be excluded by the use of reliable, commercially available tests. Hepatitis E is frequently found in AIH patients and should be considered (van Gerven 2016). The exclusion of other hepatotropic viruses such as cytomegalovirus, Epstein-Barr and herpes may only be required in cases suspicious of such infections or if the diagnosis of AIH based on the above-mentioned criteria remains inconclusive.
The probability of AIH usually decreases whenever signs of bile duct involvement are present, such as elevation of alkaline phosphatase, histological signs of cholangiopathy and detection of AMA. If one or more components of the scoring system are not evaluated, only a probable diagnosis can be made (Table 1).
Epidemiology and clinical presentation
Based on limited epidemiological data, the prevalence is estimated to range between 20 to 50 cases per million among the Caucasian population in Western Europe and North America (Jepsen 2015). The prevalence of AIH is similar to that of systemic lupus erythematosus, primary biliary cholangitis and myasthenia gravis, which also have an autoimmune aetiology (Nishioka 1997, Nishioka 1998). Among the Caucasian population in North American and Western European, AIH accounts for up to 20% of cases with chronic hepatitis (Cancado 2000). However, chronic viral hepatitis remains the major cause of chronic hepatitis in most Western societies.
Autoimmune hepatitis is part of the syndrome of chronic hepatitis, which is characterised by sustained hepatocellular inflammation for at least six months and an elevation of ALT and AST of 1.5 times the upper limit of normal. In about 49% of AIH patients an acute onset of AIH is observed and rare cases of fulminant AIH have been reported. In most cases, however, the clinical presentation is not spectacular and is characterised by fatigue, right upper quadrant pain, jaundice and occasionally also by palmar erythema and spider naevi. In later stages, the consequences of portal hypertension dominate, including ascites, bleeding oesophageal varices and encephalopathy. A specific feature of AIH is the association of extrahepatic immune-mediated syndromes including autoimmune thyroiditis, vitiligo, alopecia, nail dystrophy, ulcerative colitis, rheumatoid arthritis, and also diabetes mellitus and glomerulonephritis.
Table 1. International criteria for the diagnosis of AIH (Alvarez 1999)| Parameter | Score |
| Gender Female Male |
+ 2 0 |
| Serum biochemistry Ratio of elevation of serum alkaline phosphatase to aminotransferase >3.0 1.5–3 <1.5 |
– 2 0 + 2 |
| Total serum globulin, γ-globulin or IgG (x upper limit of normal) >2.0 1.5–2.0 1.0–1.5 <1.0 |
+ 3 + 2 + 1 0 |
| Autoantibodies (titres by immunfluorescence on rodent tissues) Adults ANA, SMA or LKM-1 >1:80 1:80 1:40 <1:40 |
+ 3 + 2 + 1 0 |
| Antimitochondrial antibody Positive Negative |
– 4 0 |
| Hepatitis viral markers Negative Positive |
+ 3 – 3 |
| History of drug use Yes No |
– 4 + 1 |
| Alcohol (average consumption) <25 gm/day >60 gm/day |
+ 2 - 2 |
| Genetic factors: HLA-DR3 or -DR4 | + 1 |
| Other autoimmune diseases | + 2 |
| Response to therapy Complete Relapse |
+ 2 + 3 |
| Liver histology Interface hepatitis Predominant lymphoplasmacytic infiltrate Rosetting of liver cells None of the above Biliary changes Other changes |
+ 3 + 1 + 1 – 5 – 3 – 3 |
| Seropositivity for other defined autoantibodies | + 2 |
Natural history and prognosis
Data describing the natural history of AIH are scarce. The last placebo-controlled immunosuppressive treatment trial containing an untreated arm was published in 1980 (Kirk 1980). The value of these studies is limited considering that these patients were only screened for then available epidemiological risk factors for viral hepatitis and were not characterised by standardised diagnostic criteria and available virological tests. Nevertheless, these studies reveal that untreated AIH had a very poor prognosis and 5- and 10-year survival rates of 50% and 10% were reported. They furthermore demonstrated that immunosuppressive treatment significantly improved survival.
Up to 30% of adult patients had histological features of cirrhosis at diagnosis. In 17% of patients with periportal hepatitis, cirrhosis developed within five years, but cirrhosis develops in 82% when bridging necrosis or necrosis of multiple lobules is present. The frequency of remission (86%) and treatment failure (14%) are comparable in patients with and without cirrhosis at presentation. Importantly, the presence of cirrhosis does not influence 10-year survival and those patients require a similarly aggressive treatment strategy (Geall 1968, Soloway 1972).
Almost half of the children with AIH already have cirrhosis at the time of diagnosis. Long-term follow-up revealed that few children can completely stop all treatment and about 70% of children receive long-term treatment (Homberg 1987, Gregorio 1997). Most of these patients relapse when treatment is discontinued, or if the dose of the immunosuppressive drug is reduced. About 15% of patients develop chronic liver failure and are transplanted before the age of 18 years.
In elderly patients, a more severe initial histological grade has been reported (Strassburg 2006). The risk of hepatocellular carcinoma varies considerably between the different diseases PBC, PSC and AIH. Particular PSC is regularly complicated by cholangiocarcinoma, gall bladder carcinoma and rarely hepatocellular carcinoma (Zenousi 2014). In contrast, occurrence of HCC in patients with AIH is a rare event and develops only in long-standing cirrhosis.
Who requires treatment?
Autoimmune hepatitis (AIH) is a remarkably treatable chronic liver disease (Manns 2001, Czaja 2010). Untreated, the prognosis of active AIH is dismal, with 5- and 10-year survival rates between 50 and 10% and a well-recognised therapeutic effect exemplified by the last placebo-controlled treatment trials (Soloway 1972, Kirk 1980). For these reasons the indication for treatment is given in any patient who has an established AIH diagnosis, elevations of aminotransferase activities (ALT, AST), an elevation of serum IgG and histological evidence of interface hepatitis or necroinflammatory activity. This has been discussed in the newest version of the AASLD (Manns 2010a) and the EASL (EASL 2015) AIH guidelines. An initial liver biopsy is recommended for confirmation of the diagnosis and for grading and staging. Biopsies are also helpful for observation of aminotransferase activities in serum reflecting inflammatory activity in the liver, which is not always closely correlated.
Who does not require treatment?
Very few patients with an established AIH diagnosis should not be treated. Rare cases, in which the initiation of standard therapy should be weighed against potential side effects, are contraindications with steroids or azathioprine, or for certain other immunosuppressants (see below). In decompensated liver cirrhosis of patients on the waiting list for liver transplantation and in individuals with complete cirrhosis and absent inflammatory activity treatment does not appear beneficial (Manns 2010a, EASL 2015).
Standard treatment strategy
Independent of the clinically- or immunoserologically-defined type of AIH, standard treatment is implemented with predniso(lo)ne alone or in combination with azathioprine. Both strategies are as effective (Manns 2001, Manns 2010a). The basic premise is based upon the findings of studies of almost three decades ago that indicated the effectiveness of steroids in AIH. Since that time, no single multicentre randomised treatment trial in AIH patients has been performed. Advances of alternative treatments are based on small cohorts and on the need to develop strategies for difficult-to-treat patients. The use of prednisone or its metabolite prednisolone, which is used more frequently in Europe, is effective since chronic liver disease does not seem to have an effect on the synthesis of prednisolone from prednisone. The exact differentiation between viral infection and autoimmune hepatitis is important. Treatment of replicative viral hepatitis with corticosteroids must be prevented as well as administration of interferon in AIH, which can lead to dramatic disease exacerbation.
Standard induction treatment and suggested follow-up examinations are summarised in Table 2. Please note the differences in preferred regimen in Europe and the US, which are delineated in the AASLD AIH Guideline (Manns 2010a). Therapy is usually administered over the course of two years. The decision between monotherapy and combination therapy is guided principally by side effects. Long-term steroid therapy leads to cushingoid side effects. Cosmetic side effects decrease patient compliance considerably (Table 3). Serious complications such as steroid diabetes, osteopenia, aseptic bone necrosis, psychiatric symptoms, hypertension and cataract formation also have to be anticipated in long-term treatment. Side effects are found in 44% of patients after 12 months and in 80% of patients after 24 months of treatment. However, predniso(lo)ne monotherapy is possible in pregnant patients. Azathioprine, on the other hand, leads to a decreased dose of prednisone. It bears a theoretical risk of teratogenicity. In addition, abdominal discomfort, nausea, cholestatic hepatitis, rash and leukopenia can be encountered. These side effects are seen in 10% of patients receiving a dose of 50 mg per day. From a general point of view, a postmenopausal woman with osteoporosis, hypertension and elevated blood glucose would be a candidate for combination therapy. In young women, pregnant women or patients with haematological abnormalities, prednisone monotherapy may be the treatment of choice.
Table 2. Treatment regimen and follow-up examinations of autoimmune hepatitis regardless of autoantibody type| Monotherapy | Combination therapy | |||||
| Prednis(ol)-one | 60 mg reduction by 10 mg/week to maintenance of 20 mg/wk reduction by 5 mg to 10 mg find lowest dose in 2.5 mg decrements | 30–60 mg reduction as in monotherapy | ||||
| Azathioprine | n.a. (maintenance with azathioprine: monotherapy: 2 mg/kg body weight) | 1 mg/kg of body weight (Europe) 50 mg (US) | ||||
| Examination | Before therapy | During therapy before remission q 4 weeks | Remission on therapy q 3–6 months | Cessation of therapy q 3 weeks (x 4) | Remission post-therapy q 3–6 months | Evaluation of relapse |
| Physical | + | + | + | + | + | |
| Liver biopsy | + | (+/-) | + | |||
| Blood count | + | + | + | + | + | |
| Aminotrans- ferases | + | + | + | + | + | + |
| Gamma glutamyl- transferase | + | + | + | |||
| Gamma- globulin | + | + | + | + | + | + |
| Bilirubin | + | + | + | + | + | + |
| Coagulation studies | + | + | + | + | + | |
| Autoanti- bodies | + | +/- | + | |||
| Thyroid function tests | + | +/- | + | |||
| Prednis(ol)one | Azathioprine |
| acne moon-shaped face striae rubra dorsal hump obesity weight gain diabetes mellitus cataracts hypertension |
nausea vomiting abdominal discomforts hepatotoxicity rash leukocytopenia teratogenicity (?) oncogenicity (?) |
One of the most important variables for treatment success is adherence. The administration of treatment is essential since most cases of relapse are the result of erratic changes of medication and/or dose. Dose reduction is aimed at finding the individually appropriate maintenance dose. Since histology lags 3 to 6 months behind the normalisation of serum parameters, therapy has to be continued beyond the normalisation of aminotransferase levels. Usually, maintenance doses of predniso(lo)ne range between 10 and 2.5 mg. After 12 to 24 months of therapy predniso(lo)ne can be tapered over the course of 4 to 6 weeks to test whether a sustained remission has been achieved. Tapering regimens aiming at withdrawal should be attempted with great caution and only after obtaining a liver biopsy that demonstrates a complete resolution of inflammatory activity. Relapse of AIH and risk of progression to fibrosis is almost universal when immunosuppression is tapered in the presence of residual histological inflammation. Withdrawal should be attempted with caution to prevent recurrence and subsequent fibrosis progression and should be discussed with the patient and closely monitored.
Outcomes of standard therapy can be classified into four categories: remission, relapse, treatment failure and stabilisation.
Remission is a complete normalisation of all inflammatory parameters including histology. The achievement of aminotransferase activities within two-fold of the upper limit of normal is not recommended as treatment goal, rather, normalisation should be aimed at. Remission is ideally the goal of all treatment regimens and ensures the best prognosis. Remission can be achieved in 65 to 75% of patients after 24 months of treatment. Remission can be sustained with azathioprine monotherapy of 2 mg/kg bodyweight (Johnson 1995). This prevents cushingoid side effects. However, side effects such as arthralgia (53%), myalgia (14%), lymphopenia (57%) and myelosuppression (6%) have been observed. Complete remission is not achieved in about 20% of patients and these patients continue to carry a risk of progressive liver injury.
Relapse is characterised by an increase in aminotransferase levels and the reccurrence of clinical symptoms either while on treatment, following tapering of steroid doses to determine the minimally required dose, or, after a complete withdrawal of therapy. Relapse happens in 50% of patients within six months of treatment withdrawal and in 80% after three years. Relapse is associated with progression to cirrhosis in 38% and liver failure in 14%. Relapse requires reinitiation of standard therapy, consideration of dosing as well as diagnosis, and perhaps a long-term maintenance dose with predniso(lo)ne or azathioprine monotherapy.
Treatment failure characterises a progression of clinical, serological and histological parameters during standard therapy. This is seen in about 10% of patients. In these cases the diagnosis of AIH has to be carefully reconsidered to exclude other etiologies of chronic hepatitis. In these patients experimental regimens can be administered or liver transplantation will become necessary.
Stabilisation is the achievement of a partial remission. Since 90% of patients reach remission within three years, the benefit of standard therapy has to be reevaluated in this subgroup of patients. Ultimately, liver transplantation provides a definitive treatment option.
Treatment of elderly patients
The presentation of acute hepatitis, clinical symptoms of jaundice, abdominal pain and malaise have a high likelihood of attracting medical attention and subsequently leading to the diagnosis of AIH (Nikias 1994). More subtle courses of AIH may not lead to clinically relevant signs and may develop unnoticed other than via routine work-up for other problems or via screening programmes. The question of disease onset in terms of initiation of immune-mediated liver disease versus the clinical consequences that become noticeable after an unknown period of disease progression is not easily resolved. Thus, “late onset” AIH may simply just reflect a less severe course of the disease with slower progression to cirrhosis. While LKM positive patients display a tendency towards an earlier presentation, both acute and subtle (earlier and late presentation) variants appear to exist in ANA positive AIH. In practice, the diagnostic dilemma is that AIH is still perceived by many as a disease of younger individuals and that therefore this differential diagnosis is less frequently considered in elderly patients with cryptogenic hepatitis or cirrhosis. Another relevant question resulting from these considerations is the issue of treatment. Standard therapy in AIH consists of steroids alone or a combination with azathioprine. In maintenance therapy azathioprine monotherapy can also be administered but induction with azathioprine alone is not effective. From a general standpoint most internists will use caution when administering steroids to elderly patients, especially in women in whom osteopenia or diabetes may be present.
Recommendations for the treatment of AIH suggest that side effects be weighed against the potential benefit of therapy, and that not all patients with AIH are good candidates for steroid treatment (Manns 2001). Controversy exists surrounding the benefit of therapy in this group of elderly patients. One cohort reported on 12 patients aged over 65 out of a total of 54 AIH patients. Cirrhosis developed after follow-up in 26% irrespective of age although the histological grade of AIH activity was more severe in the elderly group. Although 42% of the patients over 65 did not receive therapy, deaths were only reported in the younger group (Newton 1997). Another cohort of 20 patients aged over 65, reported a longer time to diagnosis (8.5 vs. 3.5 months) with patients presenting mainly with jaundice and acute onset AIH but that they showed a comparable response rate to immunosuppression to that of younger patients (Schramm 2001). The authors also noted that the prevalence of the HLA A1-B8 allotype was less frequent in older patients suggesting a role for immunogenetics.
This point was further elaborated by a report analysing 47 patients with ANA positive AIH aged 60 years and older, as well as 31 patients aged 30 years and younger in whom DR4+/DR3– prevalence was 47% (older) versus 13% (younger) patients (Czaja 2006). Steroid responsiveness was better in the older patients, in line with previous findings in the same cohort (Czaja 1993). Cirrhosis and extrahepatic immune-mediated syndromes including thyroid and rheumatologic disease (47% vs. 26%) were more prevalent in older AIH patients. However, although more treatment failures were observed in the younger patients (24% vs 5%), the rates of remission, sustained remission and relapse were similar. Interestingly, an assessment of age-stratified prevalence showed an increase after the age of 40 from 15% to over 20%.
From all this data, AIH in elderly patients appears to be characterised by a distinct clinical feature, a distinct immunogenetic profile, favourable response rates and higher rates of cirrhosis present at diagnosis, all of which contribute to the heterogeneity of AIH. A UK cohort of 164 AIH patients included 43 individuals aged 60 years (Al-Chalabi 2006). The different age groups showed no significant differences regarding serum biochemistry, autoantibody titres, time to establishment of diagnosis, and mode of presentation. The authors provided a substratification of patients below and above 40 years of age and reported that older patients had a higher median histological stage and a comparable median grade but that younger patients had more median relapse episodes and a higher median stage at follow-up biopsy. The most distinguishing clinical sign was a higher prevalence of ascites in the older group. However, rates of complete, partial and failed response were similar, and the median number of relapses was higher in younger patients, which nevertheless did not lead to differences in liver-related deaths in either group (12% vs. 15%). In comparison to the study of ANA positive AIH patients from the US (Czaja 2006), the differing findings regarding HLA association are noteworthy. In the UK study there was no differential distribution of HLA DR3 and DR4 and this questions the suggested hypothesis of a primary influence of immunogenetics on the observed clinical distinctions. The reasons for the clinical differences of AIH in older and younger patients are unclear. They may include differences in hepatic blood flow and alterations involving the regulation of cellular immunity during ageing (Talor 1991, Prelog 2006). In summary, these data suggest that AIH in elderly patients should be considered and treated (Strassburg 2006).
Alternative treatments
When standard treatment fails or drug intolerance occurs, alternative therapies such as cyclosporine, tacrolimus, cyclophosphamide, mycophenolate mofetil, rapamycin, UDCA, and budesonide can be considered (Table 4). The efficacy of most of these options has not yet been definitively decided and is only reported in small case studies.
Budesonide
Budesonide is a synthetic steroid with high first-pass metabolism in the liver, in principle with limited systemic side effects compared to conventional steroids. In comparison to prednisone the absolute bioavailability of budesonide is less than 6-fold lower (Thalen 1979) but it has an almost 90% first-pass metabolism in the liver, a higher affinity to the glucocorticoid receptor, acts as an anti-inflammatory and immunosuppressive drug and leads to inactive metabolites (6-OH-budesonide, 16-OH-prednisolone). In a pilot study treating 13 AIH patients with budesonide over a period of 9 months the drug was well-tolerated and aminotransferase levels were normalised (Danielson 1994). However, in a second study budesonide therapy was associated with a low frequency of remission and high occurrence of side effects (Czaja 2000) in 10 patients who had previously been treated with azathioprine and steroids and had not reached a satisfactory remission. This study concluded that budesonide was not a good treatment option in those patients. A third study reported that remission was induced with budesonide combination therapy in 12 previously untreated patients (Wiegand 2005). The authors performed kinetic analyses and reported that the area under the curve (AUC) of budesonide was increased in those with high inflammatory activity and cirrhosis. This finding plausibly demonstrates that in patients with portosystemic shunts in portal hypertension the effect of high hepatic first-pass metabolism that would limit typical steroid side effects is reduced.
Table 4. Alternative drugs in autoimmune hepatitis| Compound | Advantage | Disadvantage |
| Budesonide | High first pass effect Immunosuppressive action Inactive metabolites |
Cirrhosis (portosystemic shunts) and side effects |
| Cyclosporine | Satisfactory experience Potent immunosuppressant Transplant immunosuppressant |
Renal toxicity |
| Tacrolimus | Potent immunosuppressant Transplant immunosuppressant |
Renal toxicity |
| Mycophenolic acid | Favourable toxicity profile Transplant immunosuppressant |
Disappointing effectiveness |
| Cyclophosphamide | Effective | Continuous therapy Hematological side effects |
The main advantage of budesonide for the future treatment of autoimmune hepatitis would therefore be to replace prednisone in long-term maintenance therapy and induction therapy to reduce steroid side effects. To this end the first multicentre placebo-controlled randomised AIH treatment trial in 3 decades was performed with a total of 207 non-cirrhotic patients from 30 centres in nine European countries and Israel (Manns 2010b). In this trial 40 mg prednisone (reduction regimen) and azathioprine was compared to 3 mg budesonide (TID initially, reduced to BID) in combination with azathioprine. The data shows that budesonide in combination with azathioprine is efficient in inducing stable remission, is superior in comparison to a standard prednisone tapering regimen beginning with 40 mg per day and leads to a substantially superior profile of steroid-specific side effects. From these data, budesonide has emerged as an alternative first line treatment strategy for non-cirrhotic patients with AIH (Manns 2010b, EASL 2015). Budesonide is licensed for the use in AIH in many countries. Effective treatment of children with budesonide has been reported (Woynarowski 2013).
Deflazacort
This alternative corticosteroid has also been studied for immunosuppression in AIH because of its feature of fewer side effects than conventional glucocorticoids. In a pilot study 15 patients with AIH type 1 were treated with deflazacort, who had been previously treated with prednisone with or without azathioprine until they reached a biochemical remission. Remission was sustained for two years of follow-up. However, the long-term role of second-generation corticosteroids to sustain remission in AIH patients with reduced treatment-related side effects requires further controlled studies (Rebollo Bernardez 1999).
Cyclosporine A
Cyclosporine A (CyA) is a lipophylic cyclic peptide of 11 residues produced by Tolypocladium inflatum that acts on calcium-dependent signaling and inhibits T cell function via the interleukin 2 gene (Strassburg 2008). Out of the alternative AIH drugs considerable experience has been reported with CyA. CyA was successfully used for AIH treatment and was well tolerated (Alvarez 1999b, Debray 1999). The principal difficulty in advocating widespread use of CyA as first line therapy relates to its toxicity profile, particularly with long-term use (increased risk of hypertension, renal insufficiency, hyperlipidaemia, hirsutism, infection, and malignancy) (Alvarez 1999b, Debray 1999, Fernandez 1999, Heneghan 2002).
Tacrolimus
Tacrolimus is a macrolide lactone compound with immunosuppressive qualities exceeding those of CyA. The mechanism of action is similar to that of CyA but it binds to a different immunophilin (Strassburg 2008). The application of tacrolimus in 21 patients treated for one year led to an improvement of aminotransferase and bilirubin levels with a minor increase in serum BUN and creatinine levels (Van Thiel 1995). In a second study with 11 steroid-refractory patients, improvement of inflammation was also observed (Aqel 2004). A recent study demonstrated the effectiveness of tacrolimus in difficult to treat patients (Than 2016). However, although tacrolimus represents a promising immunosuppressive candidate drug, larger randomised trials are required to assess its role in the therapy of AIH.
Mycophenolic acid
Mycophenolate is a noncompetitive inhibitor of inosine monophosphate dehydrogenase, which blocks the rate-limiting enzymatic step in de novo purine synthesis and is widely used in solid organ transplantation. Mycophenolate has a selective action on lymphocyte activation, with marked reduction of both T and B lymphocyte proliferation. In a pilot study, seven patients with AIH type 1 who either did not tolerate azathioprine or did not respond to standard therapy with a complete normalisation of aminotransferase levels, were treated with mycophenolate in addition to steroids. Normalisation of aminotransferase levels was achieved in five out of seven patients within three months. These preliminary data suggested that mycophenolate may represent a promising treatment strategy for AIH (Richardson 2000). However, in a retrospective study, there was no statistically significant benefit for mycophenolate treatment in 37 patients with AIH and azathioprine failure or intolerance who were treated with mycophenolate (Hennes 2008a). Less than 50% reached remission and in the azathioprine non-responders failure was 75%. Mycophenolate has been demonstrated to be most effective as a second line therapy in patients found to be intolerant to azathioprine. There is some evidence that mycophenolate can be used as first line therapy (Zachou 2016). There is limited data available on the use of mTOR inhibitors such as everolimus in AIH (Ytting 2015).
Cyclophosphamide
The induction of remission with 1–1.5 mg per kg per day of cyclophosphamide in combination with steroids has been reported. However, the dependency of continued application of cyclophosphamide with its potentially severe haematological side effects renders it a highly experimental treatment option (Kanzler 1996).
Anti-TNF α antibodies
There is some emerging evidence that anti-TNF antibodies are capable of inducing remission in AIH patients in whom standard or alternative therapeutic options have been exhausted (Efe 2010, Umekita 2011, Weiler-Norman 2013). However, the development of AIH has also been observed under treatment with anti-TNF antibodies (Ramos-Casals 2008). Future studies will have to define the role of this therapeutic option in difficult-to-treat cases of AIH.
Ursodeoxycholic acid
Ursodeoxycholic acid is a hydrophilic bile acid with putative immunomodulatory capabilities. It is presumed to alter HLA class I antigen expression on cellular surfaces and to suppress immunoglobulin production. Uncontrolled trials have shown a reduction in histological abnormalities, clinical and biochemical improvement but not a reduction of fibrosis in four patients with AIH type 1 (Calmus 1990, Nakamura 1998, Czaja 1999). However, its role in AIH therapy or in combination with immunosuppressive therapy is still unclear.
Other alternative treatment strategies include methotrexate, anti-TNF α antibodies, and rituximab, but there is currently insufficient data on any of these.
Overlap syndromes and treatment
Overlap syndrome describes a disease condition that is not completely defined (Strassburg 2006). A valid definition is difficult (Boberg 2011). It is characterised by the coexistence of clinical, biochemical or serological features of autoimmune hepatitis (AIH), primary biliary cholangitis (PBC), primary sclerosing cholangitis (PSC), and depending on the definition, also viral hepatitis C (HCV) (Ben-Ari 1993, Colombato 1994, Duclos-Vallee 1995, Chazouilleres 1998, Angulo 2001, Rust 2008). In adult patients an overlap of PBC and AIH is most frequently encountered although it is unclear whether this is true co-existence of both diseases or an immunoserological overlap characterised by the presence of antinuclear (ANA) as well as antimitochondrial (AMA) antibodies (Poupon 2006, Gossard 2007, Silveira 2007, Al-Chalabi 2008). In many AMA negative patients with a cholestatic liver enzyme profile ANA are present. This has been termed autoimmune cholangiopathy or AMA negative PBC (Michieletti 1994).
Apart from coexisting, autoimmune liver diseases can also develop into each other, i.e., the sequential manifestation of PBC and autoimmune hepatitis. The true coexistence of AIH and PSC has only been conclusively shown in paediatric patients (Gregorio 2001). It can be hypothesised whether a general predisposition toward liver autoimmunity exists which has a cholestatic, a hepatitic and a bile duct facet, which may be variable depending upon unknown host factors. The diagnosis of an overlap syndrome relies on the biochemical profile (either cholestatic with elevated alkaline phosphatase, gamma glutamyltransferase and bilirubin, or hepatitic with elevated aspartate aminotransferase and alanine aminotransferase levels in addition to elevated gamma globulins), the histology showing portal inflammation with or without the involvement of bile ducts, and the autoantibody profile showing AMA or autoantibodies associated primarily with AIH such as liver-kidney microsomal antibodies (LKM), soluble liver antigen antibodies (SLA/LP) or ANA. In cholestatic cases cholangiography detects sclerosing cholangitis. In an overlap syndrome the classical appearance of the individual disease component is mixed with features of another autoimmune liver disease. Immunoglobulins are usually elevated in all autoimmune liver diseases.
Regarding a therapeutic strategy, the leading disease component is treated. In an overlap syndrome presenting as hepatitis, immunosuppression with prednisone (or combination therapy with azathioprine) is initiated. In cholestatic disease ursodeoxycholic acid is administered. Both treatments can be combined when biochemistry and histology suggest a relevant additional disease component (Chazouilleres 1998). Validated therapeutic guidelines for overlap syndromes are not available. It is important to realise that treatment failure in AIH may be related to an incorrect diagnosis or an overlap syndrome of autoimmune liver diseases (Potthoff 2007). Several studies show that treatment of the AIH component of overlap syndromes is important to avoid progression to cirrhosis (Chazouilleres 2006, Gossard 2007, Silveira 2007, Al-Chalabi 2008).
Liver transplantation
In approximately 10% of AIH patients liver transplantation remains the only life-saving option (Strassburg 2004). The indication for liver transplantation in AIH is similar to that in other chronic liver diseases and includes clinical deterioration, development of cirrhosis, bleeding oesophageal varices and coagulation abnormalities despite adequate immunosuppressive therapy (Neuberger 1984, Sanchez-Urdazpal 1991, Ahmed 1997, Prados 1998, Tillmann 1999, Vogel 2004). There is no single indicator or predictor for the necessity of liver transplantation. Candidates for liver transplant are usually patients who do not reach remission within four years of continuous therapy. Indicators of a high mortality associated with liver failure are histological evidence of multilobular necrosis and progressive hyperbilirubinaemia. In Europe, 4% of liver transplants are for AIH (Strassburg 2009). The long-term results of liver transplantation for AIH are excellent. The five-year survival is up to 92% (Sanchez-Urdazpal 1991, Prados 1998, Ratziu 1999) and well within the range of other indications for liver transplantation. The European liver transplant database indicates 76% survival in five years and 66% survival after 10 years (1647 liver transplantations between 1988 and 2007). When these numbers are considered it is necessary to realise that patients undergoing liver transplantation usually fail standard therapy and may therefore have a reduced life expectancy after liver transplant compared to those who achieve stable complete remission on drug therapy.
Recurrence and de novo AIH after liver transplantation
The potential of AIH to recur after liver transplantation is beyond serious debate (Schreuder 2009). The first case of recurrent AIH after liver transplant was reported in 1984 (Neuberger 1984) and was based upon serum biochemistry, biopsy findings and steroid reduction. Studies published over the years indicate that the rate of recurrence of AIH ranges between 10–35%, and that the risk of AIH recurrence is perhaps as high as 68% after five years of follow-up (Wright 1992, Devlin 1995, Götz 1999, Milkiewicz 1999, Manns 2000, Vogel 2004). It is important to consider the criteria upon which the diagnosis of recurrent AIH is based. When transaminitis is chosen as a practical selection parameter many patients with mild histological evidence of recurrent AIH may be missed. It is therefore suggested that all patients with suspected recurrence of autoimmune hepatitis receive a liver biopsy, biochemical analyses of aminotransferases as well as a determination of immunoglobulins and autoantibody titres (Vogel 2004). Significant risk factors for the recurrence of AIH have not yet been identified although it appears that the presence of fulminant hepatic failure before transplantation protects against the development of recurrent disease. Risk factors under discussion include steroid withdrawal, tacrolimus versus cyclosporine, HLA mismatch, HLA type, and LKM-1 autoantibodies. An attractive risk factor for the development of recurrent AIH is the presence of specific HLA antigens that may predispose toward a more severe immunoreactivity. In two studies recurrence of AIH appeared to occur more frequently in HLA DR3 positive patients receiving HLA DR3 negative grafts. However, this association was not confirmed in all studies. There have not been conclusive data to support the hypothesis that a specific immunosuppressive regimen represents a risk factor for the development of recurrent AIH (Gautam 2006). However, data indicate that patients transplanted for AIH require continued steroids in 64% versus 17% of patients receiving liver transplants for other conditions (Milkiewicz 1999).
Based on these results and other studies it would appear that maintenance of steroid medication in AIH patients is indicated to prevent not only cellular rejection but also graft-threatening recurrence of AIH (Vogel 2004). Steroid withdrawal should therefore be performed only with great caution. The recurrence of AIH is an important factor for the probability of graft loss. Apart from HCV and primary sclerosing cholangitis a recent report found AIH recurrence to represent the third most common reason for graft loss (Rowe 2008). Transplanted patients therefore require a close follow-up and possibly an immunosuppressive regimen including steroids, although this is controversial and not backed by prospective studies (Campsen 2008).
In addition to AIH recurrence the development of de novo autoimmune hepatitis after liver transplantation has been reported (Kerkar 1998, Jones 1999a, Salcedo 2002). The pathophysiology of this is also elusive. From a treatment point of view de novo autoimmune hepatitis, which appears to occur mostly in patients transplanted with PBC but may just be the serendipitous occurrence of AIH, is responsive to steroid treatment (Salcedo 2002).
References
Ahmed M, Mutimer D, Hathaway M, Hubscher S, McMaster P, Elias E. Liver transplantation for autoimmune hepatitis: a 12-year experience. Transplant Proc 1997;29:496.
Al-Chalabi T, Boccato S, Portmann BC, McFarlane IG, Heneghan MA. Autoimmune hepatitis (AIH) in the elderly: a systematic retrospective analysis of a large group of consecutive patients with definite AIH followed at a tertiary referral centre. J Hepatol 2006;45(4):575-83.
Al-Chalabi T, Portmann BC, Bernal W, McFarlane IG, Heneghan MA. Autoimmune hepatitis overlap syndromes: an evaluation of treatment response, long-term outcome and survival. Aliment Pharmacol Ther 2008;28(2):209-20.
Alvarez F, Berg PA, Bianchi FB, et al. International Autoimmune Hepatitis Group Report: review of criteria for diagnosis of autoimmune hepatitis. J Hepatology 1999;31:929-38.
Alvarez F, Ciocca M, Canero-Velasco C, et al. Short-term cyclosporine induces a remission of autoimmune hepatitis in children. J Hepatol 1999;30:222-7.
Angulo P, El-Amin O, Carpenter HA, Lindor KD. Development of autoimmune hepatitis in the setting of long-standing primary biliary cirrhosis. Am J Gastroenterol 2001;96(10):3021-7.
Aqel BA, Machicao V, Rosser B, Satyanarayana R, Harnois DM, Dickson RC. Efficacy of tacrolimus in the treatment of steroid refractory autoimmune hepatitis. J Clin Gastroenterol 2004;38(9):805-9.
Ben-Ari Z, Dhillon AP, Sherlock S. Autoimmune cholangiopathy: part of the spectrum of autoimmune chronic active hepatitis. Hepatology 1993;18(1):10-5.
Boberg KM, Chapman RW, Hirschfield GM, Lohse AW, Manns MP, Schrumpf E. Overlap syndromes: the International Autoimmune Hepatitis Group (IAIHG) position statement on a controversial issue. J Hepatol 2011;54(2):374-85.
Calmus Y, Gane P, Rouger P, Poupon R. Hepatic expression of class I and class II major histocompatibility complex molecules in primary biliary cirrhosis: effect of ursodeoxycholic acid. Hepatology 1990;11(1):12-5.
Campsen J, Zimmerman MA, Trotter JF, et al. Liver transplantation for autoimmune hepatitis and the success of aggressive corticosteroid withdrawal. Liver Transpl 2008;14(9):1281-6.
Cancado ELR, Porta G. Autoimmune hepatitis in South America. Dordrecht, Boston, London, Kluwer Academic Publishers, 2000.
Chazouilleres O, Wendum D, Serfaty L, Montembault S, Rosmorduc O, Poupon R. Primary biliary cirrhosis-autoimmune hepatitis overlap syndrome: clinical features and response to therapy. Hepatology 1998;28(2):296-301.
Chazouilleres O, Wendum D, Serfaty L, Rosmorduc O, Poupon R. Long term outcome and response to therapy of primary biliary cirrhosis- autoimmune hepatitis overlap syndrome. J Hepatol 2006;44(2):400-6.
Colombato LA, Alvarez F, Cote J, Huet PM. Autoimmune cholangiopathy: the result of consecutive primary biliary cirrhosis and autoimmune hepatitis? Gastroenterology 1994;107(6):1839-43.
Czaja AJ, Carpenter HA, Lindor KD. Ursodeoxycholic acid as adjunctive therapy for problematic type 1 autoimmune hepatitis: a randomized placebo-controlled treatment trial. Hepatology 1999;30(6):1381-6.
Czaja AJ, Carpenter HA, Santrach PJ, Moore SB. Significance of HLA DR4 in type 1 autoimmune hepatitis. Gastroenterology 1993;105:1502-7.
Czaja AJ, Carpenter HA. Distinctive clinical phenotype and treatment outcome of type 1 autoimmune hepatitis in the elderly. Hepatology 2006;43(3):532-8.
Czaja AJ, Lindor KD. Failure of budesonide in a pilot study of treatment-dependent autoimmune hepatitis. Gastroenterology 2000;119(5):1312-6.
Czaja AJ, Manns MP. Advances in the diagnosis, pathogenesis, and management of autoimmune hepatitis. Gastroenterology 2010;139(1):58- 72 e4.
Danielson A, Prytz H. Oral budesonide for treatment of autoimmune chronic hepatitis. Aliment Pharmacol Ther 1994;8:585-90.
Debray D, Maggiore G, Giradet JP, Mallet E, Bernard O. Efficacy of cyclosporin A in children with type 2 autoimmune hepatits. J Pediatr 1999;135:111-4.
Devlin J, Donaldson P, Portman B, et al. Recurrence of autoimmune hepatitis following liver transplantation. Liver Transpl Surg 1995;1:162-5.
Dienes HP, Popper H, Manns M, Baumann W, Thoenes W, Meyer zum Büschenfelde K-H. Histologic features in autoimmune hepatitis. Z Gastroenterol 1989;27:327-30.
Duclos-Vallee JC, Hadengue A, Ganne-Carrie N, et al. Primary biliary cirrhosis-autoimmune hepatitis overlap syndrome. Corticoresistance and effective treatment by cyclosporine A. Dig Dis Sci 1995;40(5):1069-73.
EASL Clinical Practice Guidelines: management of cholestatic liver diseases. J Hepatol 2009;51(2):237-67. EASL Clinical Practice Guidelines: Autoimmune hepatitis. J Hepatol. 2015;63(4):971-1004.
Efe C, Purnak T, Ozaslan E. Anti TNF-alpha therapy can be a novel treatment option in patients with autoimmune hepatitis. Aliment Pharmacol Ther 2010;32(1):115; author reply 6-7.
Fernandez NF, Redeker AG, Vierling JM, Villamil FG, Fong T-L. Cyclosporine therapy in patients with steroid resistant autoimmune hepatitis. Am J Gastroenterol 1999;94:241-8.
Gautam M, Cheruvattath R, Balan V. Recurrence of autoimmune liver disease after liver transplantation: a systematic review. Liver Transpl 2006;12(12):1813-24.
Geall MG, Schoenfield LJ, Summerskill WHJ. Classification and treatment of chronic active liver disease. Gastroenterology 1968;55:724-9.
Gelpi C SE, Rodriguez-Sanchez JL. Autoantibodies against a serine tRNA-protein complex implicated in cotranslational selenocysteine insertion. Proc Natl Acad Sci U S A 1992;89(20):9739-43.
Gossard AA, Lindor KD. Development of autoimmune hepatitis in primary biliary cirrhosis. Liver Int 2007;27(8):1086-90.
Götz G, Neuhaus R, Bechstein WO, et al. Recurrence of autoimmune hepatitis after liver transplantation. Transplant Proc 1999;31:430-1.
Gregorio GV, Portman B, Reid F, et al. Autoimmune hepatitis in childhood: a 20-year experience. Hepatology 1997;25:541-7.
Gregorio GV, Portmann B, Karani J, et al. Autoimmune hepatitis/sclerosing cholangitis overlap syndrome in childhood: a 16-year prospective study. Hepatology 2001;33(3):544-53.
Heneghan MA, McFarlane IG. Current and novel immunosuppressive therapy for autoimmune hepatitis. Hepatology 2002;35(1):7-13.
Hennes EM, Oo YH, Schramm C, et al. Mycophenolate mofetil as second line therapy in autoimmune hepatitis? Am J Gastroenterol. 2008;103(12):3063-70.
Hennes EM, Zeniya M, Czaja AJ, et al. Simplified criteria for the diagnosis of autoimmune hepatitis. Hepatology 2008;48(1):169-76.
Jepsen P, Gronbaek L, Vilstrup H. Worldwide Incidence of Autoimmune Liver Disease. Dig Dis. 2015;33 Suppl 2:2-12.
Johnson PJ, McFarlane IG, Williams R. Azathioprine for long-term maintenance of remission in autoimmune hepatitis. N Engl J Med. 1995;333(15):958-63.
Johnson PJ, McFarlane IG. Meeting report: International autoimmune hepatitis group. Hepatology 1993;18:998-1005.
Jones DE, James OF, Portmann B, Burt AD, Williams R, Hudson M. Development of autoimmune hepatitis following liver transplantation for primary biliary cirrhosis. Hepatology 1999;30(1):53-7.
Kanzler S, Gerken G, Dienes HP, Meyer zum Büschenfelde K-H, Lohse AW. Cyclophosphamide as alternative immunosuppressive therapy for autoimmune hepatitis - report of three cases. Z Gastroenterol 1996;35:571-8.
Kerkar N, Hadzic N, Davies ET, et al. De-novo autoimmune hepatitis after liver transplantation. Lancet 1998;351(9100):409-13.
Kirk AP, Jain S, Pocock S, Thomas HC, Sherlock S. Late results of the Royal Free Hospital prospective controlled trial of prednisolone therapy in hepatitis B surface antigen negative chronic active hepatitis. Gut 1980;21:7893.
Mackay IR, Taft LI, Cowling DC. Lupoid hepatitis. Lancet 1956;2:1323-6.
Manns M, Gerken G, Kyriatsoulis A, Staritz M, Meyer zum Büschenfelde KH. Characterisation of a new subgroup of autoimmune chronic active hepatitis by autoantibodies against a soluble liver antigen. Lancet 1987;1(8528):292-4.
Manns MP, Bahr MJ. Recurrent autoimmune hepatitis after liver transplantation - when non-self becomes self. Hepatology 2000;32(4):868-70.
Manns MP, Czaja AJ, Gorham JD, et al. Diagnosis and management of autoimmune hepatitis. Hepatology 2010;51(6):2193-213.
Manns MP, Griffin KJ, Sullivan KF, Johnson EF. LKM-1 autoantibodies recognize a short linear sequence in P450IID6, a cytochrome P-450 monooxygenase. J Clin Invest 1991;88:1370-8.
Manns MP, Johnson EF, Griffin KJ, Tan EM, Sullivan KF. Major antigen of liver kidney microsomal antibodies in idiopathic autoimmune hepatitis is cytochrome P450db1. J Clin Invest 1989;83:1066-72.
Manns MP, Strassburg CP. Autoimmune hepatitis: clinical challenges. Gastroenterology 2001;120(6):1502-17.
Manns MP, Woynarowski M, Kreisel W, et al. Budesonide induces remission more effectively than prednisone in a controlled trial of patients with autoimmune hepatitis. Gastroenterology 2010;139(4):1198-206.
Michieletti P, Wanless IR, Katz A, et al. Antimitochondrial antibody negative primary biliary cirrhosis: a distinct syndrome of autoimmune cholangitis. Gut 1994;35(2):260-5.
Milkiewicz P, Hubscher SG, Skiba G, Hathaway M, Elias E. Recurrence of autoimmune hepatitis after liver transplantation. Transplantation 1999;68(2):253-6.
Nakamura K, Yoneda M, Yokohama S, et al. Efficacy of ursodeoxycholic acid in Japanese patients with type 1 autoimmune hepatitis [see comments]. J Gastroenterol Hepatol 1998;13(5):490-5.
Neuberger J, Portmann B, Calne R, Williams R. Recurrence of autoimmune chronic active hepatitis following orthotopic liver grafting. Transplantation 1984;37(4):363-5.
Newton JL, Burt AD, Park JB, Mathew J, Bassendine MF, James OF. Autoimmune hepatitis in older patients. Age Ageing 1997;26(6):441-4.
Nikias GA, Batts KP, Czaja AJ. The nature and prognostic implications of autoimmune hepatitis with acute presentation. J Hepatol. 1994;21:866-71.
Nishioka M, Morshed SA, Kono K, et al. Frequency and significance of antibodies to P450IID6 protein in Japanese patients with chronic hepatiis C. J Hepatol 1997;26:992-1000.
Nishioka M, Morshed SA, McFarlane IG. Geographical variation in the frequency and characteristics of autoimmune liver diseases. Amsterdam: Elsevier; 1998.
Potthoff A, Deterding K, Trautwein C, et al. Steroid treatment for severe acute cryptogenic hepatitis. Z Gastroenterol 2007;45(1):15-9.
Poupon R, Chazouilleres O, Corpechot C, Chrétien Y. Development of autoimmune hepatitis in patients with typical primary biliary cirrhosis. Hepatology. 2006;44(1):85-90.
Prados E, Cuervas-Mons V, De La Mata M, et al. Outcome of autoimmune hepatitis after liver transplantation. Transplantation 1998;66:1645-50.
Prelog M. Aging of the immune system: a risk factor for autoimmunity? Autoimmun Rev 2006;5(2):136-9.
Ramos-Casals M, Brito-Zeron P, Soto MJ, Cuadrado MJ, Khamashta MA. Autoimmune diseases induced by TNF-targeted therapies. Best Pract Res Clin Rheumatol 2008;22(5):847-61.
Ratziu V, Samuel D, Sebagh M, et al. Long-term follow-up after liver transplantation for autoimmune hepatitis: evidence of recurrence of primary disease. J Hepatol 1999;30(1):131-41.
Rebollo Bernardez J, Cifuentes Mimoso C, Pinar Moreno A, et al. Deflazacort for long-term maintenance of remission in type I autoimmune hepatitis. Rev Esp Enferm Dig 1999;91(9):630-8.
Richardson PD, James PD, Ryder SD. Mycophenolate mofetil for maintenance of remission in autoimmune hepatitis in patients resistant to or intolerant of azathioprine. J Hepatol 2000;33(3):371-5.
Rowe IA, Webb K, Gunson BK, Mehta N, Haque S, Neuberger J. The impact of disease recurrence on graft survival following liver transplantation: a single centre experience. Transpl Int 2008;21(5):459-65.
Rust C, Beuers U. Overlap syndromes among autoimmune liver diseases. World J Gastroenterol 2008;14(21):3368-73.
Salcedo M, Vaquero J, Banares R, et al. Response to steroids in de novo autoimmune hepatitis after liver transplantation. Hepatology 2002;35(2):349-56.
Sanchez-Urdazpal L, Czaja AJ, Van Holk B. Prognostic features and role of liver transplantation in severe corticoid-treated autoimmune chronic active hepatitis. Hepatology 1991;15:215-21.
Schramm C, Kanzler S, zum Büschenfelde KH, Galle PR, Lohse AW. Autoimmune hepatitis in the elderly. Am J Gastroenterol 2001;96(5):1587-91.
Schreuder TC, Hubscher SG, Neuberger J. Autoimmune liver diseases and recurrence after orthotopic liver transplantation: what have we learned so far? Transpl Int 2009;22(2):144-52.
Silveira MG, Talwalkar JA, Angulo P, Lindor KD. Overlap of autoimmune hepatitis and primary biliary cirrhosis: long-term outcomes. Am J Gastroenterol 2007;102(6):1244-50.
Soloway RD, Summerskill WH, Baggenstoss AH, et al. Clinical, biochemical, and histological remission of severe chronic active liver disease: a controlled study of treatments and early prognosis. Gastroenterology 1972;63(5):820-33.
Stechemesser E, Klein R, Berg PA. Characterization and clinical relevance of liver-pancreas antibodies in autoimmune hepatitis. Hepatology 1993;18:1-9.
Strassburg CP, Bahr MJ, Becker T, Klempnauer J, Manns MP. Progress in immunosuppression. Chirurg 2008;79(2):149-56.
Strassburg CP, Becker T, Klempnauer J, Manns MP. Liver transplantation: deciding between need and donor allocation. Internist (Berl) 2004;45(11):1233-45.
Strassburg CP, Manns MP. Liver transplantation: indications and results. Internist (Berl) 2009.
Strassburg CP, Manns MP. Primary biliary liver cirrhosis and overlap syndrome. Diagnosis and therapy. Internist (Berl) 2004;45(1):16-26. Strassburg CP, Manns MP. Autoantibodies and autoantigens in autoimmune hepatitis. Semin Liver Dis 2002;22(4):339-52.
Strassburg CP, Manns MP. Autoimmune hepatitis in the elderly: what is the difference? J Hepatol 2006;45(4):480-2. Strassburg CP, Manns MP. Autoimmune hepatitis versus viral hepatitis C. Liver 1995;15:225-32.
Strassburg CP, Obermayer-Straub P, Alex B, et al. Autoantibodies against glucuronosyltransferases differ between viral hepatitis and autoimmune hepatitis. Gastroenterology 1996;111(6):1576-86.
Strassburg CP, Obermayer-Straub P, Manns MP. Autoimmunity in liver diseases. Clin Rev Allergy Immunol 2000;18(2):127-39. Strassburg CP. Autoimmune liver diseases and their overlap syndromes. Praxis (Bern 1994) 2006;95:1363-81.
Stravitz RT, Lefkowitch JH, Fontana RJ, et al. Autoimmune acute liver failure: proposed clinical and histological criteria. Hepatology 2011;53(2):517-26.
Talor E, Rose NR. Hypothesis: the aging paradox and autoimmune disease. Autoimmunity 1991;8(3):245-9.
Thalen A, Brattsand R. Synthesis and anti-inflammatory properties of budesonide, a new non-halogenated glucocorticoid with high local activity. Arzneimittelforschung 1979;29(11):1687-90.
Than NN, Wiegard C, Weiler-Normann C, et al. Long-term follow-up of patients with difficult to treat type 1 autoimmune hepatitis on Tacrolimus therapy. Scand J Gastroenterol. 2016;51(3):329-36.
Tillmann HL, Jackel E, Manns MP. Liver transplantation in autoimmune liver disease-selection of patients. Hepatogastroenterology 1999;46(30):3053-9.
Umekita K, Miyauchi S, Ueno S, et al. Improvement of rheumatoid arthritis and autoimmune hepatitis in a patient treated with the tumour necrosis factor inhibitor, etanercept. Intern Med 2011;50(11):1245-9.
Van Gerven NM, van der Eijk AA, Pas SD, et al. Seroprevalence of Hepatitis E Virus in Autoimmune Hepatitis Patients in the Netherlands. J Gastrointestin Liver Dis. 2016;25(1):9-13.
Van Thiel DH, Wright H, Carroll P, et al. Tacrolimus: A potential new treatment for autoimmune chronic active hepatitis: results of an open- label preliminary trial. Am J Gastroenterol 1995;90:771-6.
Vogel A, Heinrich E, Bahr MJ, et al. Long-term outcome of liver transplantation for autoimmune hepatitis. Clin Transplant 2004;18(1):62-9.
Volkmann M, Luithle D, Zentgraf H, et al. SLA/LP/tRNP((Ser)Sec) antigen in autoimmune hepatitis: identification of the native protein in human hepatic cell extract. J Autoimmun 2010;34(1):59-65.
Volkmann M, Martin L, Baurle A, et al. Soluble liver antigen: isolation of a 35-kd recombinant protein (SLA-p35) specifically recognizing sera from patients with autoimmune hepatitis. Hepatology 2001;33(3):591-6.
Waldenström J. Leber, Blutproteine und Nahrungseiweisse. Dtsch Gesellsch Verd Stoffw 1950;15:113-9.
Weiler-Normann C, Schramm C, Quaas A, et al. Infliximab as a rescue treatment in difficult-to-treat autoimmune hepatitis. J Hepatol. 2013;58(3):529-34.
Wiegand J, Schuler A, Kanzler S, et al. Budesonide in previously untreated autoimmune hepatitis. Liver Int 2005;25(5):927-34.
Wies I, Brunner S, Henninger J, et al. Identification of target antigen for SLA/LP autoantibodies in autoimmune hepatitis [see comments]. Lancet 2000;355(9214):1510-5.
Woynarowski M, Nemeth A, Baruch Y, et al. Budesonide versus prednisone with azathioprine for the treatment of autoimmune hepatitis in children and adolescents. The Journal of paediatrics 2013;163(5):1347-53.e1.
Wright HL, Bou-Abboud CF, Hassanenstein T, et al. Disease recurrence and rejection following liver transplantation for autoimmune chronic active liver disease. Transplantation 1992;53:136-9.
Ytting H, Larsen FS. Everolimus treatment for patients with autoimmune hepatitis and poor response to standard therapy and drug alternatives in use. Scand J Gastroenterol. 2015;50(8):1025-31.
Zachou K, Gatselis NK, Arvaniti P, et al. A real-world study focused on the long-term efficacy of mycophenolate mofetil as first-line treatment of autoimmune hepatitis. Aliment Pharmacol Ther. 2016;43(10):1035-47.
Zenouzi R, Weismuller TJ, Hubener P, et al. Low risk of hepatocellular carcinoma in patients with primary sclerosing cholangitis with cirrhosis. Clinical gastroenterology and hepatology: the official clinical practice journal of the American Gastroenterological Association 2014;12(10):1733-8.
